N-terminal Amino Acid Sequencing of Mouse Monoclonal Antibodies Using the Edman Method
N-terminal structural analysis is the first step in protein structural analysis. This analysis employs the Edman method (sequential cleaving of amino acids from the N-terminal of the protein to determine the amino acid sequence), which is the most reliable method for determining amino acid sequences.
The PPSQ-31A/33A Protein Sequencer System automates the Edman reaction, HPLC separation and detection, and data analysis to determine the amino acid sequence from the N-terminal.
The N-terminal amino acid sequencing of mouse monoclonal antibodies using the Edman method is introduced below.
Test and research flow
1. Subject mouse monoclonal antibodies (60 pmol) to SDS-PAGE (under reducing conditions).
2. After electrophoresis, conduct electroblotting onto a PVDF membrane and stain with CBB.
3. Excise the circled regions in Fig. 1 and analyze them by PPSQ-31A.

Fig. 1 PVDF Membrane
Fig. 2 shows the analysis results for the light chain down to Cycle 5.
The Cycle 1 results indicate that the N-terminal amino acid residue is D (aspartic acid). The Cycle 2 results indicate that the second amino acid group from the N-terminal is V (valine). Analysis to the 21st residue reveals the sequence from the N-terminal to be: Asp-Val-Val-Met-Thr-Gln-Thr-Pro-Leu-Thr-Leu-Ser-Val-Thr-Ile-Gly-Gln-Pro-Ala-Ser-Ile.
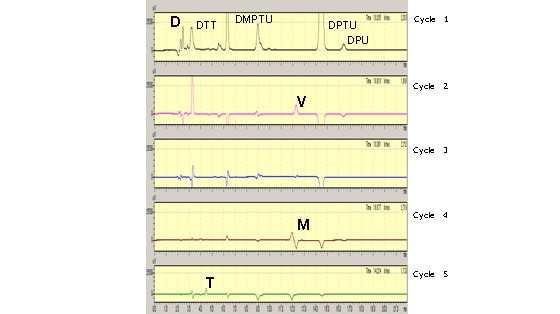
Fig. 2 Light Chain Results (Cycle 2 and Beyond Are Differential Chromatograms)
Fig. 3 shows the analysis results for the heavy chain down to Cycle 5.
The PTH amino acid derivative peak is not expressed, indicating that the N-terminal amino group (α amino group) of the heavy chain is modified.
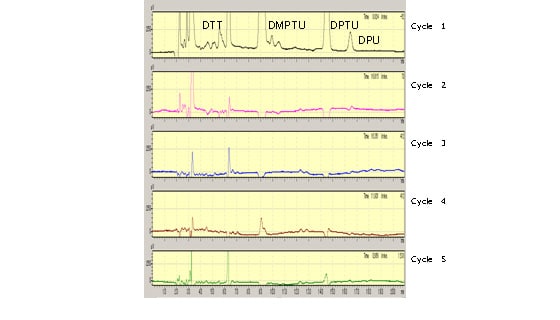
Fig. 3 Heavy Chain Results (Cycle 2 and Beyond Are Differential Chromatograms)
Protein Sequencer

Features:
The PPSQ-51A/53A continues the tradition of providing reliable and sensitive N-terminal protein sequencing to researchers through automated Edman Degradation. Whether transitioning over from another sequencer or adopting the technique for the first time, the intuitive software and robust Shimadzu hardware will make protein sequencing easy on your laboratory.


